Instead of picking a single model, Ensemble Method combines multiple models in a certain way to fit the training data. Here are the two primary ways: "bagging" and "boosting." In "bagging", we take a subset of training data (pick n random sample out of N training data, with replacement) to train up each model. After multiple models are trained, we use a voting scheme to predict future data.
Random Forest is one of the most popular bagging models; in addition to selecting n training data out of N at each decision node of the tree, it randomly selects m input features from the total M input features (m ~ M^0.5). Then it learns a decision tree from that. Finally, each tree in the forest votes for the result. Here is the R code to use Random Forest:
> library(randomForest)
#Train 100 trees, random selected attributes
> model <- randomForest(Species~., data=iristrain, nTree=500)
#Predict using the forest
> prediction <- predict(model, newdata=iristest, type='class')
> table(prediction, iristest$Species)
> importance(model)
MeanDecreaseGini
Sepal.Length 7.807602
Sepal.Width 1.677239
Petal.Length 31.145822
Petal.Width 38.617223
"Boosting" is another approach in Ensemble Method. Instead of sampling the input features, it samples the training data records. It puts more emphasis, though, on the training data that is wrongly predicted in previous iterations. Initially, each training data is equally weighted. At each iteration, the data that is wrongly classified will have its weight increased.
Gradient Boosting Method is one of the most popular boosting methods. It is based on incrementally adding a function that fits the residuals.
Set i = 0 at the beginning, and repeat until convergence.
- Learn a function Fi(X) to predict Y. Basically, find F that minimizes the expected(L(F(X) – Y)), where L is the lost function of the residual
- Learning another function gi(X) to predict the gradient of the above function
- Update Fi+1 = Fi + a.gi(X), where a is the learning rate
Below is Gradient-Boosted Tree using the decision tree as the learning model F. Here is the sample code in R:
> library(gbm)
> iris2 <- iris
> newcol = data.frame(isVersicolor=(iris2$Species=='versicolor'))
> iris2 <- cbind(iris2, newcol)
> iris2[45:55,]
Sepal.Length Sepal.Width Petal.Length Petal.Width Species isVersicolor
45 5.1 3.8 1.9 0.4 setosa FALSE
46 4.8 3.0 1.4 0.3 setosa FALSE
47 5.1 3.8 1.6 0.2 setosa FALSE
48 4.6 3.2 1.4 0.2 setosa FALSE
49 5.3 3.7 1.5 0.2 setosa FALSE
50 5.0 3.3 1.4 0.2 setosa FALSE
51 7.0 3.2 4.7 1.4 versicolor TRUE
52 6.4 3.2 4.5 1.5 versicolor TRUE
53 6.9 3.1 4.9 1.5 versicolor TRUE
54 5.5 2.3 4.0 1.3 versicolor TRUE
55 6.5 2.8 4.6 1.5 versicolor TRUE
> formula <- isVersicolor ~ Sepal.Length + Sepal.Width + Petal.Length + Petal.Width
> model <- gbm(formula, data=iris2, n.trees=1000, interaction.depth=2, distribution="bernoulli")
Iter TrainDeviance ValidDeviance StepSize Improve
1 1.2714 -1.#IND 0.0010 0.0008
2 1.2705 -1.#IND 0.0010 0.0004
3 1.2688 -1.#IND 0.0010 0.0007
4 1.2671 -1.#IND 0.0010 0.0008
5 1.2655 -1.#IND 0.0010 0.0008
6 1.2639 -1.#IND 0.0010 0.0007
7 1.2621 -1.#IND 0.0010 0.0008
8 1.2614 -1.#IND 0.0010 0.0003
9 1.2597 -1.#IND 0.0010 0.0008
10 1.2580 -1.#IND 0.0010 0.0008
100 1.1295 -1.#IND 0.0010 0.0008
200 1.0090 -1.#IND 0.0010 0.0005
300 0.9089 -1.#IND 0.0010 0.0005
400 0.8241 -1.#IND 0.0010 0.0004
500 0.7513 -1.#IND 0.0010 0.0004
600 0.6853 -1.#IND 0.0010 0.0003
700 0.6266 -1.#IND 0.0010 0.0003
800 0.5755 -1.#IND 0.0010 0.0002
900 0.5302 -1.#IND 0.0010 0.0002
1000 0.4901 -1.#IND 0.0010 0.0002
> prediction <- predict.gbm(model, iris2[45:55,], type="response", n.trees=1000)
> round(prediction, 3)
[1] 0.127 0.131 0.127 0.127 0.127 0.127 0.687 0.688 0.572 0.734 0.722
> summary(model)
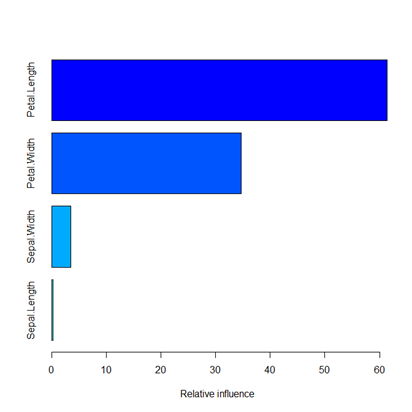
var rel.inf
1 Petal.Length 61.4203761582
2 Petal.Width 34.7557511871
3 Sepal.Width 3.5407662531
4 Sepal.Length 0.2831064016
The GBM R package also gave the relative importance of the input features, as shown in the bar graph.



{{ parent.title || parent.header.title}}
{{ parent.tldr }}
{{ parent.linkDescription }}
{{ parent.urlSource.name }}